|
This section of the Alignment Details page displays data for each of the BLAST or BLAT high-scoring pairs (HSPs) and shows a pairwise alignment report from the BLAST or BLAT result.
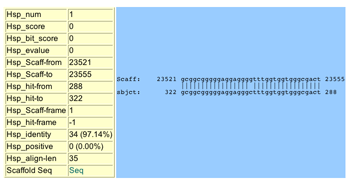
Depending on the tool (i.e. BLAST), this data might include:
- Hsp_num The high-scoring pair number, within
the alignment.
- Hsp_score The score for the HSP.
- Hsp_bit_score The bit score for the HSP.
- Hsp_evalue The E-value for the HSP.
- Hsp_Scaff-from The number of the first base in the alignment along the scaffold.
- Hsp_Scaff-to The number of the last base in the alignment along the scaffold.
- Hsp_hit-from The number of the first base in the alignment along the hit sequence.
- Hsp_hit-to The number of the last base in the alignment along the hit sequence.
- Hsp_Scaff-frame The reading frame of the scaffold.
- Hsp_Hit-frame The reading frame of the hit sequence.
- Hsp_identity The number (and percentage) of identical bases or amino acids in the alignment
- Hsp_positive The number (and percentage) of positive, or conservative, matches (not including identities) in the alignment.
- Hsp_align-len The alignment length of the HSP.
- Scaffold Seq A link to the scaffold genomic sequence for the HSP.
|