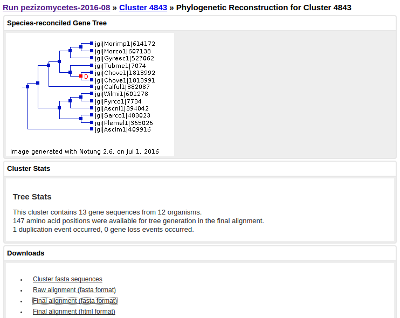

For those MCL clustering runs associated with nodes on the Mycocosm tree, species trees are also computed using single-copy orthologs in combination with RAxML, FastTree, and manual curation of the resulting trees.
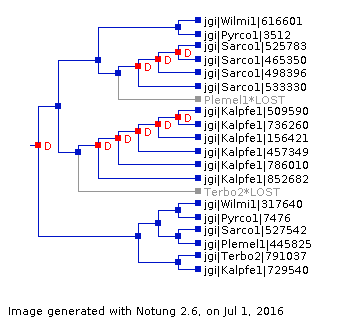
Gene trees are computed with FastTree and show the phylogenetic relationship between the proteins in a cluster.
For each cluster, the protein sequences are aligned using the MAFFT multiple sequence alignment program. Well-aligned regions are then extracted using Gblocks. Trees are then computed with FastTree. The image shown is generated using Notung (although the tree itself is computed with FastTree). Notung is also used to infer gene duplications and losses. Notice that in the tree, several genes are indicated as duplicated by a red letter 'D' at the node where the duplication occurred, and that two genes lost are noted in grey. Notung infers this by reconciling the gene tree with the species tree computed for the Mycocosm node.

Statistics on how many genes, organisms, amino acid positions were used to build the gene tree. Statistics on gene gain and loss, reported by Notung, are also given.
Download files are provided, including