Using KEGG, you can browse through both metabolic and regulatory pathways. The metabolic pathways are displayed initially. To browse through the enzyme nomenclature, follow theEnzyme Commission Numbers link. This takes you to Kyoto University'sEnzyme EC Numbers site.
Quick Tips
- To view regulatory pathways, clickKEGG Regulatory Pathways below the page header.
- To return to viewing metabolic pathways, clickKEGG Metabolic Pathways below the page header.
From the KEGG entry page, you can view a list of enzymes involved in a given reference pathway, or you can retrieve a list of JGI-predicted genes (gene models) involved in the pathway.
Metabolic Pathways Summary Table
The summary table on the home page lists the number of models available in each pathway by model set (see below).
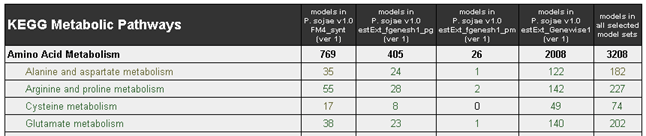
- The bold count lines represent the sum of models per pathway, for each model set.
- If you selected multiple gene model sets, an additional column at the end of the table "Models in all selected model sets" shows a summary of models per pathway, for each model set.
- To see a list ofenzymes involved in each pathway, click the pathway name, or click theModels in all selected model setsnumber for that pathway.
- To see the list of gene models (predicted genes related to a pathway), click a number in a "models in" column.

