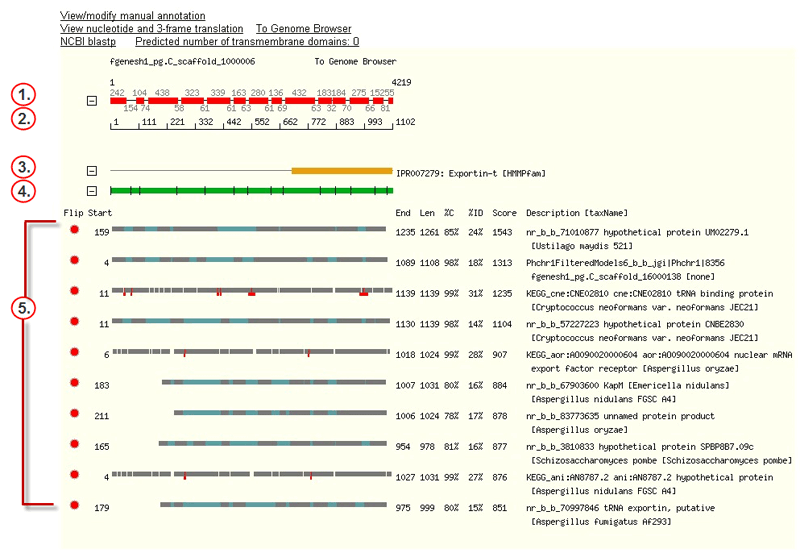

The hit tracks on the Protein page include the following features:
![]() The JGI-predicted gene (plus strand), showing lengths in base pairs of predicted exons (red rectangles) and introns (black lines).Blue rectangles mark untranslated regions, or UTRs (noncoding exons). The numbers immediately above the track are exon lengths; those below, intron lengths. The first and last bases are numbered in the upper left and upper right. To view the nucleotide sequence, click the gene to open theGene Sequence Viewer.
The JGI-predicted gene (plus strand), showing lengths in base pairs of predicted exons (red rectangles) and introns (black lines).Blue rectangles mark untranslated regions, or UTRs (noncoding exons). The numbers immediately above the track are exon lengths; those below, intron lengths. The first and last bases are numbered in the upper left and upper right. To view the nucleotide sequence, click the gene to open theGene Sequence Viewer.
![]() A scale showing the numbering of amino acid residues across the length of the protein sequence derived from the gene.
A scale showing the numbering of amino acid residues across the length of the protein sequence derived from the gene.
![]() Domain structures in the InterPro database that have alignments (orange rectangles) with the translated gene. To collapse these into one additive track (showing alignments with any of the previously displayed InterPro entities), click the- to the left of the first InterPro track. Thetext for each track links to the European Bioinformatics Institute's InterPro web page for records with that InterPro ID. The text displays the InterPro ID number, a description of the InterPro entity, and in brackets, the algorithm used to determine the match.
Domain structures in the InterPro database that have alignments (orange rectangles) with the translated gene. To collapse these into one additive track (showing alignments with any of the previously displayed InterPro entities), click the- to the left of the first InterPro track. Thetext for each track links to the European Bioinformatics Institute's InterPro web page for records with that InterPro ID. The text displays the InterPro ID number, a description of the InterPro entity, and in brackets, the algorithm used to determine the match.
![]() The protein sequence derived from the predicted gene, withblack lines marking splice junctions. To view the entire amino acid sequence in FASTA format, click anywhere on thegreen track.
The protein sequence derived from the predicted gene, withblack lines marking splice junctions. To view the entire amino acid sequence in FASTA format, click anywhere on thegreen track.
 Alignments (grey rectangles) of proteins from external databases with the translated gene. TheProtein Alignment Tracks are active areas that can be used to retrieve alignment details, switch to a flipped view, or link to external web sites.
Alignments (grey rectangles) of proteins from external databases with the translated gene. TheProtein Alignment Tracks are active areas that can be used to retrieve alignment details, switch to a flipped view, or link to external web sites.