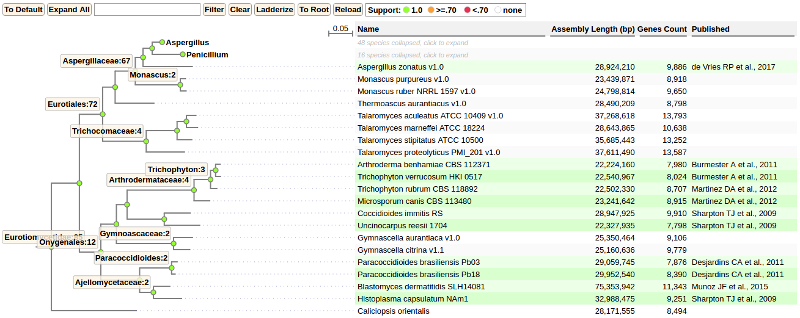

Tree view represented by two major parts: tree and table. Tree side represents annotated species tree created using the tools described above. Representation of the tree side is standard. Distance to parent and support value both represented here. Distance shown as a length of the link to the parent node while support as color label based on scale. Support value scale has following labels:
Exact values of distance to the parent and support could be viewed on tree node selection panel when given node is selected.
Table (right side of the screen) shows list of individual species in tabular form. Columns shows
Each node on the tree could be highlighted by hover over the node with mouse. Following operations are available on any highlighted node:
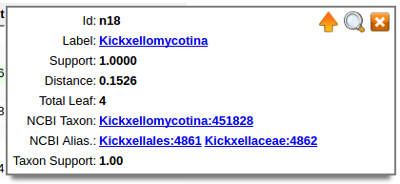
Selected node panel always appears on top right corned of the browser window. It appears when any node is selected. It has following information:
 move current tree "root" one level up
move current tree "root" one level up make currently selected node a new "root" for the tree
make currently selected node a new "root" for the tree clear selection and hide selected node panel
clear selection and hide selected node panelWe annotate our species tree with NCBI taxonomy labels using "best bidirectional match with cutoff" approach. For each node we compute list of it leaf nodes (with known NCBI taxonomy) and than look on NCBI taxonomy tree for the node that has most of those nodes. It could be that we have "perfect" match - all of leaf nodes belong to one or more NCBI nodes. Or in some cases it is "less than perfect"" match, but still the best across all possible pair of ours and NCBI nodes and ratio of "matched" leaf nodes to "total" leaf nodes is grater than a cutoff value. This ratio become "Taxon Support" value. The top-most NCBI node become an assigned NCBI taxon for given node on our tree. If we have more than one best matches from the same hierarchy - all lower taxons become Taxon Aliases, we basically cannot say on what level of NCBI taxonomy between all aliases our node must be placed.
We do our best to assign valid and meaningful NCBI labels to nodes on our tree, however, as expected, majority of nodes on our tree does not have NCBI labels.
On the top of the tree view there are several important controls.
This button will restore 'default' expand/collapse state of the tree. Usually that mean some nodes are expanded and some are collapsed to give you the 'best' visual representation of the tree.
This button will expand all nodes on the tree. Some trees could be big.
This control let you search across nodes. You can enter some text and nodes will be searched across all tree for the match. The text could be, for example, species name, or NCBI taxon. Filtered result shows all nodes that match selection and hide or 'deem' all other nodes. Clear button removes filtering, but does not change expand/collapse state of the tree.
Use of this control will cycle over 4 possible modes of tree 'ladderization'. This will change the order of species/strand rows on table view. Ladderization mean soring children nodes on each tree node by certain criteria. 2 modes related to how we sort nodes: (1) by total distance to the root; (2) by sum of all distances to the root for nodes below given. Results will vary depending of specific tree. This two modes may be ascending ore descending giving total 4 possible options.
This button will move current 'virtual' tree root to the 'actual'tree root. Usually that mean you will see whole Fungi tree.
This button will reload tree data structure from the underlying storage.