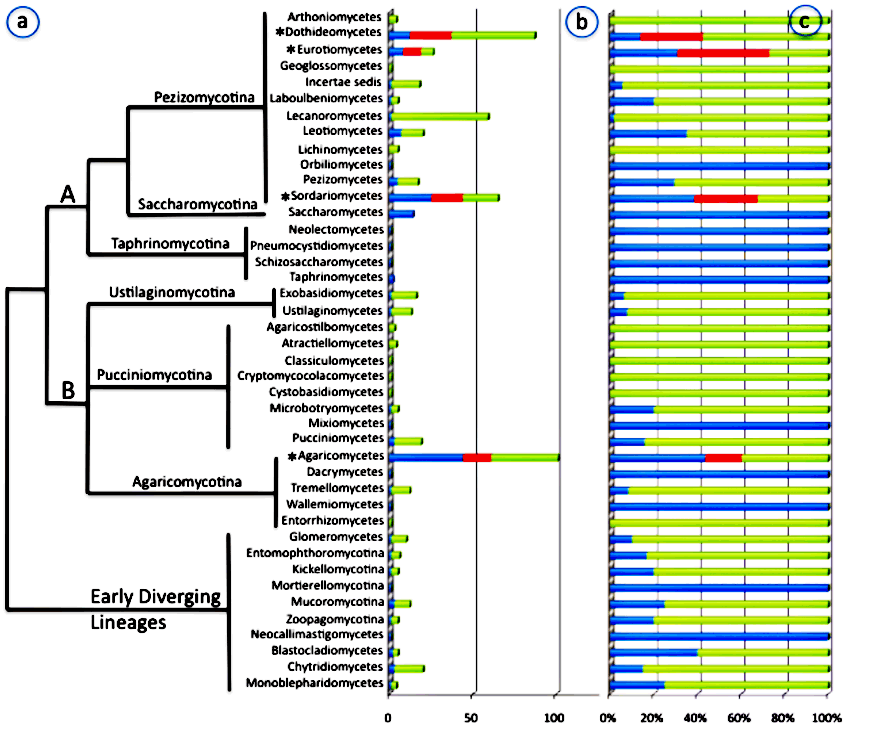
Family level sampling of fungal genomes across the Fungal Tree of Life.
Blue = completed or in progress, Red = proposed for Tier One sampling, Green = remaining unsampled families.
A=Ascomycota, B=Basidiomycota.
*The four classes represent the most phylogenetically diverse classes of nonlichenized fungi will be Tier One targets for sequencing.
With an estimated 1.5 million species, Fungi represent one of the largest branches of the Tree of Life. They have an enormous impact on human affairs and ecosystem functioning, owing to their diverse activities as decomposers, pathogens, and mutualistic symbionts. And perhaps more than any other group of nonphotosynthetic organisms, fungi are essential biological components of the global carbon cycle. Collectively, they are capable of degrading almost any naturally occurring biopolymer and numerous human-made ones. As such, fungi hold considerable promise in the development of alternative fuels, carbon sequestration and bioremediation of contaminated ecosystems.
The use of fungi for the continued benefit of humankind, however, requires an accurate understanding of how they interact in natural and synthetic communities. The ability to sample environments for complex fungal metagenomes is rapidly becoming a reality and will play an important part in harnessing fungi for industrial, energy and climate management purposes. However, our ability to accurately analyze these data relies on well-characterized, foundational reference data of fungal genomes.
To bridge this gap in our understanding of fungal diversity, an international research team in collaboration with the Joint Genome Institute of the Department of Energy has embarked on a five-year project to sequence 1000 fungal genomes from across the Fungal Tree of Life. The team comprises Joseph Spatafora (Oregon State University), Jason Stajich (University of California at Riverside), Kevin McCluskey (Fungal Genetics Stock Center), Pedro Crous (Centraal Bureau voor Schimmelcultures, Netherlands), Gillian Turgeon (Cornell University), Daniel Lindner (USDA Forest Service), Kerry O'Donnell and Todd Ward (USDA ARS), Antonis Rokas (Vanderbilt University), Louise Glass (University of California at Berkeley), Betsy Arnold (University of Arizona), Francis Martin (INRA, France) and Igor Grigoriev (JGI DOE). The overall plan is to fill in gaps in the Fungal Tree of Life by sequencing at least two reference genomes from the more than 500 recognized families of Fungi. In doing so, this project has the core goal of providing reference information to inform research on plant-microbe interactions, microbial emission and capture of greenhouse gasses, and environmental metagenomic sequencing.
1000 Fungal Genomes Community website