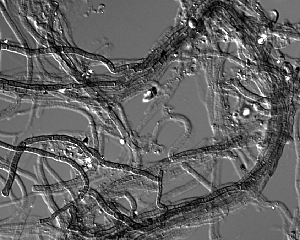
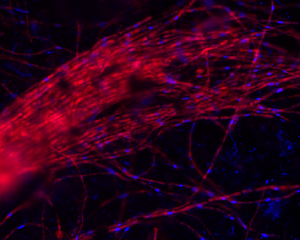
Note: there are two assemblies available for this organism. This portal contains the assembly of metagenomic sequencing of a streamer community. There is also an assembly of DNA obtained from a pure culture.
Acidomyces richmondensis is an acidophilic filamentous fungus belonging to the Dothideomycetes. A. richmondensis can be found growing at and above pH 0.7 in subterranean acid mine drainage in the Richmond Mine at Iron Mountain, California. Surveys of phylogenetic marker genes and fluorescent in situ hybridization microscopy have confirmed that a variety of Eukaryotes, including protists and filamentous fungi, co-occur with a mix of acidophilic Bacteria, Archaea, and Viruses that form both streamer and floating biofilm communities (Baker et al. 2004; Baker et al. 2009). These acid mine drainage biofilms serve as excellent model systems for detailed study of microbial communities via a variety of culture-independent molecular methods (Denef et al. 2010). Streamer biofilms occur in high-flow portions of acid mine drainage, and are dominated by filamentous fungal communities of reduced complexity. The genomic sequencing of an isolate of A. richmondensis, as well as the de novo assembly of a near-complete fungal genome from metagenomic sequencing of a streamer community, serve as starting points for understanding fungal metabolism at acidic pH, the role these fungi play in the larger microbial community, and the population metagenomics of filamentous fungi in a well-characterized natural model system.
Fungi from extreme environments may also express glycosyl hydrolases and other enzymes that are secreted extracellularly and that may have unique properties relevant to industrial applications. Acid mine drainage biofilms exist as polysaccharide-rich matrices that contain significant amounts of cellulose (Jiao et al. 2010). Genomic characterization of microbial communities containing A. richmondensis have revealed a variety of novel putative fungal glycosyl hydrolases, and transcriptomic and proteomic studies have confirmed the in situ expression of some of these enzymes. Secreted A. richmondensis enzymes have adapted to the extremely acidic (pH<1), metal-rich (≈200 mM Fe), and thermophilic (40-50 °C) environment of acid mine drainage. These glycosyl hydrolases may prove useful as components of acid-stable enzyme cocktails for deconstruction of biomass feedstocks for bioenergy production. More generally, comparative genomics of A. richmondensis will help reveal the adaptations that allow for enzyme stability and activity under such extreme conditions.
Genome Reference(s)
Mosier AC, Miller CS, Frischkorn KR, Ohm RA, Li Z, LaButti K, Lapidus A, Lipzen A, Chen C, Johnson J, Lindquist EA, Pan C, Hettich RL, Grigoriev IV, Singer SW, Banfield JF
Fungi Contribute Critical but Spatially Varying Roles in Nitrogen and Carbon Cycling in Acid Mine Drainage.
Front Microbiol. 2016;7():238. doi: 10.3389/fmicb.2016.00238
Baker, B., M. Lutz, S. Dawson, P. Bond and J. Banfield (2004). "Metabolically active eukaryotic communities in extremely acidic mine drainage." Applied and Environmental Microbiology 70(10): 6264-6271.
Baker, B., G. Tyson, L. Goosherst and J. Banfield (2009). "Insights into the Diversity of Eukaryotes in Acid Mine Drainage Biofilm Communities." Applied and Environmental Microbiology 75(7): 2192.
Denef, V. J., R. S. Mueller and J. F. Banfield (2010). "AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature." ISME J 4(5): 599-610.
Jiao, Y., G. D. Cody, A. K. Harding, P. Wilmes, M. Schrenk, K. E. Wheeler, J. F. Banfield and M. P. Thelen (2010). "Characterization of extracellular polymeric substances from acidophilic microbial biofilms." Appl Environ Microbiol 76(9): 2916-22.
