Image Credit: Stephane Hacquard
This genome was sequenced as part of the 1000 Fungal Genomes Project - Deep Sequencing of Ecologically-relevant Dikarya, and more specifically as part of the Endophyte Genome Sequencing project, which seeks to sequence members of diverse lineages of endophytic species found in Arabidopsis, Populus and other plants to examine the functional diversity of fungi with a shared evolutionary history.
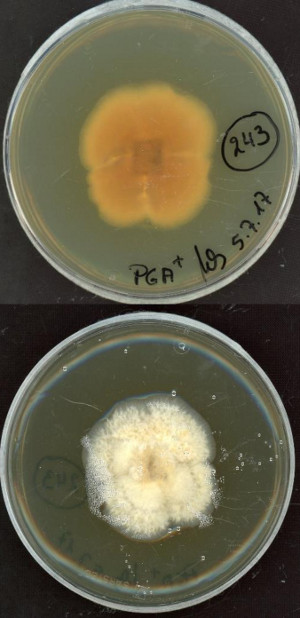
The genus Dendryphion (Ascomycota phylum, Dothideomycetes class, Pleosporales order) comprises fungi that have not been extensively described yet. Some species have been reported to colonize the roots of healthy plants. The sequenced Dendryphion nanum MPI-CAGE-CH-0243 strain has been isolated from surface sterilized roots of the flowering plant Cardamine hirsuta (a close relative of Arabidopsis thaliana) grown in the Cologne Agricultural Soil (CAS). The sequencing of this fungal isolate is part of a larger project aiming at sequencing the genomes of numerous phylogenetically diverse root-associated fungi from Arabidopsis, Populus, and other plant hosts for further comparative genome analysis. Unravelling the genomic signatures reflecting the adaptation of these microbes to the host cell environment represent a promising way to better understand how the endophytic lifestyle evolved in phylogenetically unrelated fungal species. In addition, comparative genome analysis with saprotrophic, mycorrhizal and pathogenic fungi will provide new insights into the strategies used by the fungus to colonize plant roots as well as its biocontrol potential.
Researchers who wish to publish analyses using data from unpublished CSP genomes are respectfully required to contact the PI and JGI to avoid potential conflicts on data use and coordinate other publications with the CSP master paper(s).
Genome Reference(s)
Mesny F, Miyauchi S, Thiergart T, Pickel B, Atanasova L, Karlsson M, Hüttel B, Barry KW, Haridas S, Chen C, Bauer D, Andreopoulos W, Pangilinan J, LaButti K, Riley R, Lipzen A, Clum A, Drula E, Henrissat B, Kohler A, Grigoriev IV, Martin FM, Hacquard S
Genetic determinants of endophytism in the Arabidopsis root mycobiome.
Nat Commun. 2021 Dec 10;12(1):7227. doi: 10.1038/s41467-021-27479-y
