|
With this search you can find JGI-predicted genes (gene models) whose translated sequences have Smith-Waterman alignments with proteins in a variety of relevant public databases. To launch this search, use only the Gene Model Hits form.
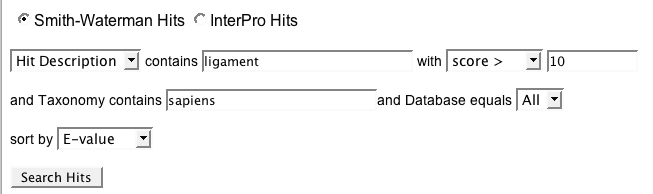
- Make sure the Smith-Waterman radio button is selected.
- From the first pull-down menu, select a field from the external database to search. The options are Hit Id, Hit Name (nearly always the same as the Hit Id), and Hit Description.
- In the contains text box, type your search term (e.g.,
20302 for a hit ID or hit name, H+ ATPase for a hit description).
- From the second pull-down menu, select the statistical criteria you would like to use to limit the results, either score or E-value.
- In the text box, type the E-value or score you want to use as the cutoff (e.g.,
900 for a score, 1e-25 for an E-value.
- In the Taxonomy contains text box, type any taxonomic term to which you want to limit results (e.g.,
oryza).
- Choose a database (or All) from the Database equals menu.
- Choose a sort option from the sort by menu. The options are Score, E-value, or Protein Id.
- Click Search Hits.
Results from Searches for Hits to Multiple Databases
|