The genome sequence and gene predictions of Monacrosporium haptotylum were not determined by the JGI, but were downloaded from NCBI and have been published (Tejashwari Meerupati et al., 2013). Please note that this copy of the genome is not maintained by the author and is therefore not automatically updated.
This species has equivalent name Dactylellina haptotyla CBS 200.50
Pezizomycotina is the largest subphylum and includes the vast majority of filamentous, fruit-body-producing species. Molecular phylogeny resolves Orbiliomycetes and Pezizomycetes as the early-diverging lineages of the Pezizomycotina, with the remaining seven classes sampled forming a well-supported crown clade. The Orbiliomycetes consists of a single order (Orbiliales) and one family (Orbiliaceae). This family is best known for containing nematode-trapping fungi.
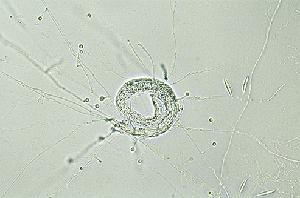
Together with A. oligospora, M. haptotylum is the nematode-trapping fungus in which the infection mechanism has been studied in most detail. A major advantage of using M. haptotylum in infection experiments is that the trap cells (knobs) can be isolated from a mycelium growing in liquid cultures. The isolated traps are functionally intact, that is, they can capture and kill nematodes, including Caenorhabditis briggsae. Accordingly, the system provides unique opportunities to identify genes that are differentially expressed in the trap cells and in the fungus during the various stages of infection.
Genome Reference(s)
Meerupati T, Andersson KM, Friman E, Kumar D, Tunlid A, Ahrén D
Genomic mechanisms accounting for the adaptation to parasitism in nematode-trapping fungi.
PLoS Genet. 2013 Nov;9(11):e1003909. doi: 10.1371/journal.pgen.1003909
Andersson KM, Meerupati T, Levander F, Friman E, Ahrén D, Tunlid A
Proteome of the nematode-trapping cells of the fungus Monacrosporium haptotylum.
Appl Environ Microbiol. 2013 Aug;79(16):4993-5004. doi: 10.1128/AEM.01390-13
Meerupati Tejashwari, Andersson Karl-Magnus, Friman Eva, Kumar Dharmendra, Tunlid Anders, Ahrén Dag
Genomic Mechanisms Accounting for the Adaptation to Parasitism in Nematode-Trapping Fungi
PLoS Genet. 2013;9(11):e1003909. doi: 10.1371/journal.pgen.1003909
