JGI portals provide protein family annotation data for our organisms and organism group pages. Protein functional regions or domains provide insight into protein's function. Protein sequences are compared to multiple sequence alignments and profile hidden Markov models (HMM) for every known protein family in the Pfam database managed by the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI). The Pfam is a database of protein sequences where families are sets of protein regions that share a significant degree of sequence similarity, thereby suggesting homology. For additional information on their methods, see their help pages here: EMBL-EBI Pfam Help Summary. Briefly, each EMBL-EBI Pfam-A entry is assigned an accession number (format = PF00123) and new sequences are annotated based on the E-value generated by the HMMER3 statistical package which equals the number of hits that would be expected to have a score equal to or better than this value by chance alone. Each Pfam-A has a unique Domain Gathering Threshold which defines inclusion for subsequent protein sequences.
EMBL-EBI classifies Pfam entries in one of six ways:
Family: A collection of related protein regions
Domain: A structural unit
Repeat: A short unit which is unstable in isolation but forms a stable structure when multiple copies are present
Motifs: A short unit found outside globular domains
Coiled-Coil: Regions that predominantly contain coiled-coil motifs, regions that typically contain alpha-helices that are coiled together in bundles of 2-7.
Disordered: Regions that are conserved, yet are either shown or predicted to contain bias sequence composition and/or are intrinsically disordered (non-globular).
There are two pages containing information about a portal's Pfam annotations. Here, on the Pfam Annotations Summary Page you can search for and create lists of Pfam annotations within comparator genomes. These annotations have been assigned by generating a Gathering Score for each protein sequence. If the gathering score for given unknown sequence is greater than EBML-EBI's Pfam-A gathering score threshold for a particular domain, that pfam domain is predicted. Selected proteins can be viewed on the Proteins Browser Page. where additional information on the genomic location, context, and gene/protein size can be found.
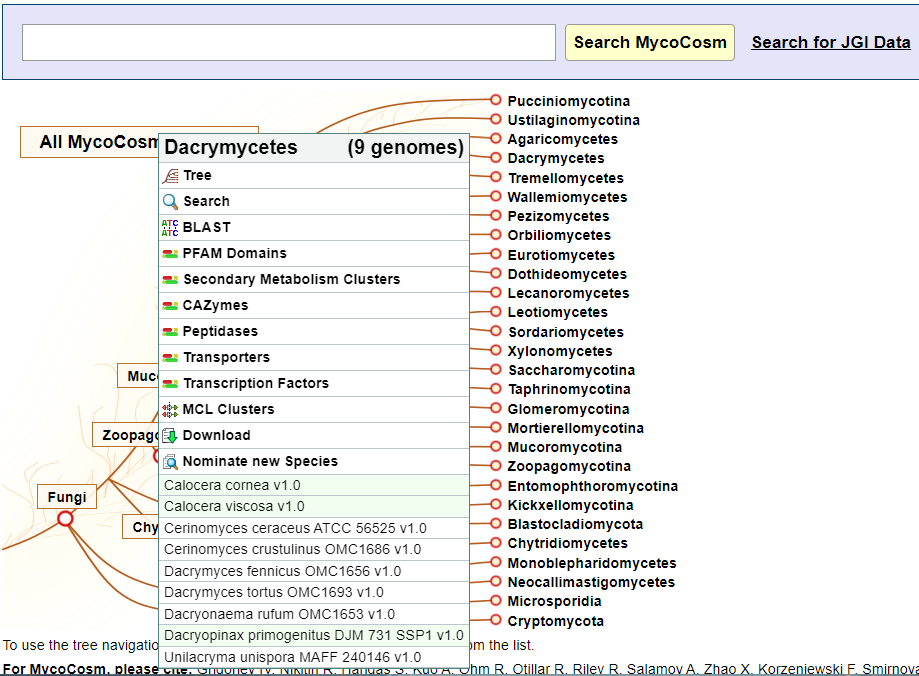
To access the Pfam Annotations Summary Page from the homepage tree, choose a group of interest and select the Pfam Domains link from the pop-up menu.

To access this page from the organism page, select the ANNOTATIONS tab and then click PFAM DOMAINS. The Proteins Browser Page. is accessed by selecting the proteins of interest from this annotations page as described below.
You are viewing a summary of all of or a subset of the Pfam Annotations in a portal. The group of annotations you are viewing is indicated here:
At the top of the page are controls to filter the displayed Transporter annotations.
By default the filter is empty and you can see all Pfam annotations available for the organism or organism group. The subset of Pfam's you are viewing is indicated above the menu bar.

 This button shows the default view for the portal's Pfam annotations when you first view the page.
This button shows the default view for the portal's Pfam annotations when you first view the page. This button shows all of the Pfam annotations. Selecting the Pfam annotation code will direct you to the specific Pfam Family page describing this Pfam-A annotation.
This button shows all of the Pfam annotations. Selecting the Pfam annotation code will direct you to the specific Pfam Family page describing this Pfam-A annotation. The blank input field is for filtering Pfam annotations. You can enter keywords like "ABC" or "HSP70" (without double quotes) to identify proteins matching the filter terms
The blank input field is for filtering Pfam annotations. You can enter keywords like "ABC" or "HSP70" (without double quotes) to identify proteins matching the filter terms Widen or narrow your search results using these drop-down menus in conjunction with your keyword(s) search.
Widen or narrow your search results using these drop-down menus in conjunction with your keyword(s) search.  After entering the filter value into the field, hit the Enter key on the keyboard or click this "Filter" button to apply your new filter and show only the matching results.
After entering the filter value into the field, hit the Enter key on the keyboard or click this "Filter" button to apply your new filter and show only the matching results. This button removes any filter and shows all Pfam annotations for the organism or organism group specified in the top right hand corner of the page..
This button removes any filter and shows all Pfam annotations for the organism or organism group specified in the top right hand corner of the page.. Checking this box allows you to hide the portion of the Pfam list without any matching results and shows only the Pfams matching the current filter. By default, this box is selected.
Checking this box allows you to hide the portion of the Pfam list without any matching results and shows only the Pfams matching the current filter. By default, this box is selected. Checking this box allows you to see the totals for only the Pfam annotations matching the filter term. If you uncheck this check box you will see a pair of totals separated by a "/" character. The left number represents the total number of annotations matching the filter criteria, and the right number shows how many annotations total there are without the filter. By default, this box is selected.
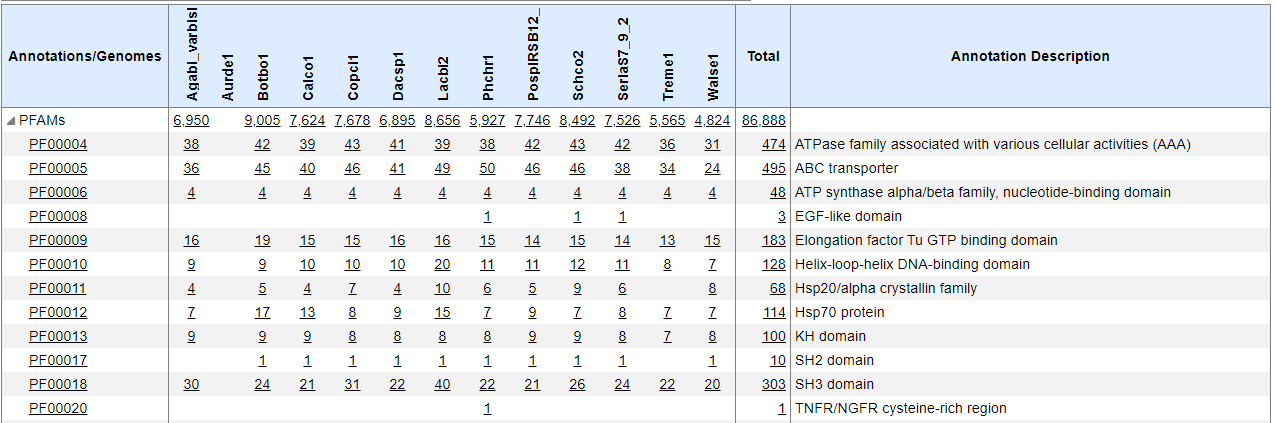
Checking this box allows you to see the totals for only the Pfam annotations matching the filter term. If you uncheck this check box you will see a pair of totals separated by a "/" character. The left number represents the total number of annotations matching the filter criteria, and the right number shows how many annotations total there are without the filter. By default, this box is selected. This table shows the Pfam hierarchy and the total number of Pfam Annotations for every organism in the group that is specified in the upper right hand corner of the page.
Every row represents one Pfam annotation or group of annotations. The first column titled Annotations/Genomes contains the Pfam-A number. Clicking on a Pfam-A number will take you to the corresponding EMBL-EBI Pfam database page for additional information.  The second set of columns each represent one organism, and the organism's name links to that organism's description page. Every cell below the organism contains the total number of proteins with a given Pfam annotation for that organism and links directly to the Protein Browser for additional information.
The second set of columns each represent one organism, and the organism's name links to that organism's description page. Every cell below the organism contains the total number of proteins with a given Pfam annotation for that organism and links directly to the Protein Browser for additional information.
Totals and sub-totals for Pfam families are shown according to the Pfam hierarchy. The PFAMs row and the Totals column contain totals across the row or column, respectively. The Annotation Description column contains a description of the Pfam identified in the Annotations/Genomes column. You can click on any value in the table to view the stated number of proteins in the Protein Browser. Here you can access additional information regarding the Pfams of interest.