|
Help Home > The Genome Browser > The Genome Viewer > Alignment Details > Alignment Graphic Display Alignment Graphic Display |
|
The alignment can be viewed as an x-y plot graphic (the default) or as a linear graphic. Both graphics show precisely how the bases in the two sequences match up. The color key (BLAST only) is the same for both versions. x-y Plot Graphic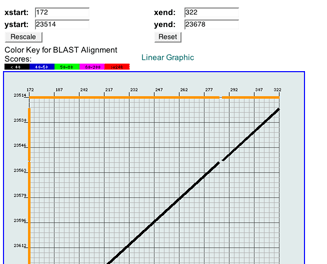
The default view on this page is the x-y Plot Graphic. In this graphic, base positions along the scaffold are marked by numbers on the vertical (y) axis, and base positions along the hit sequence are marked by numbers on the horizontal (x) axis. The alignment is represented by one or more plotted segments, each representing a high-scoring pair (HSP), colored according to the alignment score color key (shown directly above the plot). You can customize the display by setting the base ranges for each sequence in the text boxes at the top and clicking Rescale.
Quick Tip
Linear GraphicTo view the alignment in a linear display, click Linear Graphic. This graphic displays base numbers for the scaffold along the top and base numbers for the hit sequence along the bottom. Each high-scoring pair (HSP) is represented by a filled-in region between the two axes that is color coded by alignment score. To jump to an alignment hit report for one of the HSPs, click the colored trapezoid representing it. Quick Tips
|