Getting Started - Tools Overview - KEGG - GO - KOG - Track Editor - Browser - BLAST - Protein Page - Annotation Page
KEGG
KEGG is the Kyoto Encyclopedia of Genes and Genomes. You can search or browse through KEGG metabolic and regulatory pathways to retrieve information about enzymes, pathways, and proteins related to JGI-predicted genes.
KEGG divides metabolism into eleven main pathways, each of which is further
divided into reference pathways. Similarly, KEGG divides regulation of
genetic information into four main pathways, which are also further divided
into reference pathways.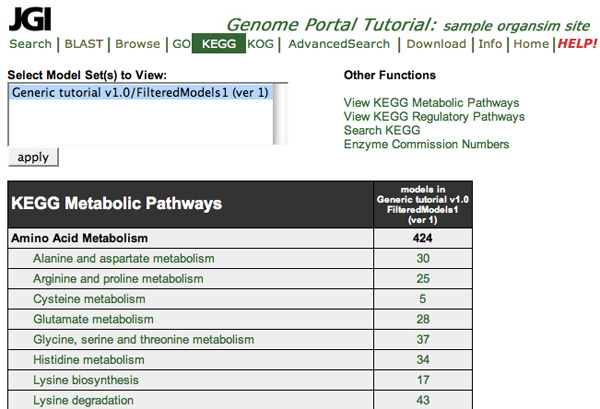
Using the KEGG browser, you can:
- view enzymes related to a pathway.
- view predicted genes related to a pathway.
- view maps of reference pathways.
- browse through KEGG pathways.
- search KEGG.
- compare models to KEGG assignments in related organisms.
View Enzymes Related to a Pathway
You can view the enzymes related to a reference pathway by clicking the corresponding Pathway link on the list of reference pathways.
The page heading shows the KEGG map number for the reference pathway and the name of the reference pathway.
The column headings in the list of enzymes are: |
|
EC Number |
The Enzyme Commission number for the enzyme, a link to a list of pathways involving that enzyme. |
Description |
A description of the enzyme's activity. |
Portal name, e.g., Postia placenta v1.0 |
The number of predicted genes in the assembly that are related to the enzyme, a link to a list of JGI-predicted genes (if any) related to the enzyme. The list is the same as a search result for the enzyme's EC number. |
- To view a diagram or rendering of the reference pathway with JGI-predicted genes highlighted in red, click the MAP number.
- To view a list of JGI-predicted genes related to one enzyme, click the number under the portal name.
- To see what other pathways involve a specific enzyme, click its EC Number.
Switching Pathways
When you click an EC Number to view the list of pathways involving an enzyme, you will see a list, including the pathway from which you selected the enzyme. The list shows the name of the main pathway ("Main Map") and the name of the reference pathway ("Sub Map").
To view the enzymes related to a different reference pathway that involves the same enzyme, click the name of the pathway in the Sub Map column.
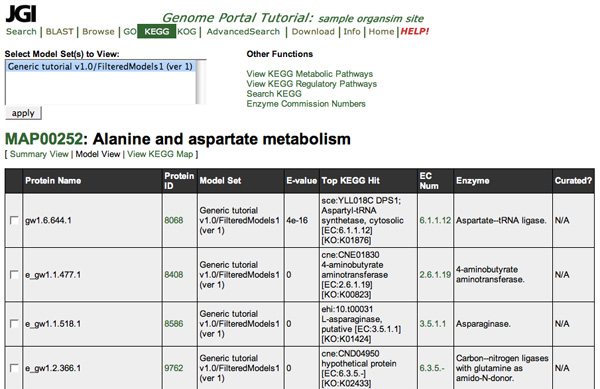
View Predicted Genes Related to a Pathway
You can view the list of genes related to a pathway by clicking the corresponding Pathway link in the list of reference pathways. Then click the Model View link under the pathway name. You can also download sequence or view multiple alignments with ClustalW.
The page heading shows the KEGG map number for the reference pathway and the name of the reference pathway.
The column headings in the list of genes are: |
|
Prot name |
JGI's name for the gene. |
Prot Id |
JGI's ID number for the gene, derived from its name. |
E-value |
The expectation value for the association of the gene with the enzyme. |
Top KEGG hit |
The protein in the KEGG database with the best BLAST alignment to the transcription of the predicted gene. The ID, name, EC number, taxonomy ID, and taxonomy name in the KEGG database are listed. Click here for a complete list of the KEGG three letter organism codes (i.e.rso) |
EC Num |
The Enzyme Commission number for the related enzyme. |
Enzyme |
A description of the related enzyme's action. |
Annotation |
The number of annotations for the gene in the genome, a link to the annotation area if it is active. |
- To view a diagram of the reference pathway, click the MAP number.
- To see the Protein page for one of the gene models, click the corresponding Prot ID.
- To go to the annotation page for a specific gene model, click its "Curated?" designation.
At the bottom of the list are buttons for retrieving FASTA sequences or ClustalW alignments.
To retrieve sequence in FASTA format:
- Click the check box next to each gene for which you want to retrieve sequence.
- Click Retrieve FASTA.
To retrieve a ClustalW alignment:
- Click the check box next to each gene that you want to include in the alignment. (You must choose more than one gene.)
- Click Clustalw.
To clear your selections, click Uncheck all.
View Maps of Reference Pathways
You can view a diagram or rendering of a reference pathway by clicking the MAP number on a list of enzymes or gene models for that pathway. In the diagrams, rectangles denote enzymes; rounded rectangles denote other reference pathways; and small circles denote compounds. Enzymes related to JGI-predicted genes are shown in red.
The map page offers links to other tools at Kyoto University. Data from these tools are not specific to the JGI-sequenced organism.
- To view results from a search of the Kyoto University link database, click LinkDB Search.
- To see information about orthologous enzymes in selected organisms, click Ortholog Table.
- To view a diagram for the same pathway in a different organism, select an organism from the menu and click Exec.
The map itself may contain links to the LinkDB for enzymes, compounds, and other reference pathways.
- To see the list of links for an enzyme, compound, or reference pathway, click its symbol on the map.
Browse through KEGG Pathways
You can browse through both metabolic and regulatory pathways in the KEGG tool. When you begin using KEGG, the metabolic pathways are displayed.
- To view regulatory pathways, click KEGG Regulatory Pathways below "other functions".
- To return to viewing metabolic pathways, click KEGG Metabolic Pathways below "other functions".
You can also browse through enzyme nomenclature by following the Enzyme Commission Numbers link. This takes you to Kyoto University's Enzyme EC Numbers site.
Search KEGG
You can search for enzymes in KEGG by Enzyme Commission (EC) number or by keyword. The results allow you to view information about predicted genes related to a given enzyme.
To search by EC number,
- Select "EC Number" from the Search Options menu.
- Type the number (e.g.,
2.6.1.1) in the EC field. - Click Submit.
To search by keyword,
- Select "Keyword" from the Search Options menu.
- Type the query word (e.g.,
gamma-lyase) in the unlabeled field to the right of the Search Options menu. - Click Submit.
The species selector is automatically set to the organism whose site you are viewing.
The KEGG search results depend on the type of search.
Using KEGG Search Results
KEGG Search results are returned as a list of enzymes that contain the keyword, or the single enzyme that matches the EC number.
| The data returned for a successful search includes: | |
EC number |
The Enzyme Commission number for the enzyme, a link to the list of genes associated with the enzyme. |
Definition |
A description of the enzyme. |
Alternative Name |
Another name for the enzyme, if one exists. |
Catalytic Activity |
The reaction catalyzed by the enzyme. |
Cofactors |
Cofactors involved in the reaction, if any. |
Associated Diseases |
Diseases associated with the enzyme, if any. |
If you search for a keyword but no matches are found, you will see only the text "Search KEGG enzyme names for keyword:" followed by your search term.
- To see a list of JGI-predicted genes associated with one enzyme listed in a keyword search result, click its EC number.
EC number search results include the list of JGI-predicted genes associated with the enzyme, if any exist in the species of interest.
The list shows: |
|
Species |
The name of the species and the assembly number in which the match occurs. |
Model Set |
Which model track this model is from. |
Protein ID |
JGI's ID number for the predicted gene, a link to the Protein page for that gene. |
Protein Name |
JGI's protein name for the predicted gene and a link to the Protein page for that gene. |
Source |
The source database used to match the enzyme with the predicted gene. |
E-value |
The expectation value for the match (the probability of a match occurring by chance alone). |
Top KEGG Hit |
The name of the protein with the best BLAST alignment to the predicted protein. |
Curated? |
Denotes whether or not this predicted gene has been manually curated. The designation is a link to the annotation page, if it is active. |
- To view the Protein page for a predicted gene, click its Protein ID or Protein Name.
- To view the annotation page for a predicted gene, click its Curated? designation.